|
Back in 2010 I tinkered around with 3D graphics, and I learned how to use OpenGL, a 3D graphics engine, with the only language I knew well, Haskell. Eager to try it out, I embarked on a project to build from scratch a molecular graphics engine, because cool images of molecules always impressed me. Below are some snapshots from the program Hmol that I wrote: Hmol is publicly available via Github under the GNU GPLv3 License (details in the Licensing section at the bottom). I distinctly remember finishing much of the development during my exam period—no classes and no homework, so lots of spare time! User Info A quick introduction to the main features, and how to try the program out. User Interface Features
Pre-built Molecules
Setting Up Download all the Hmol source files from Github and install the following:
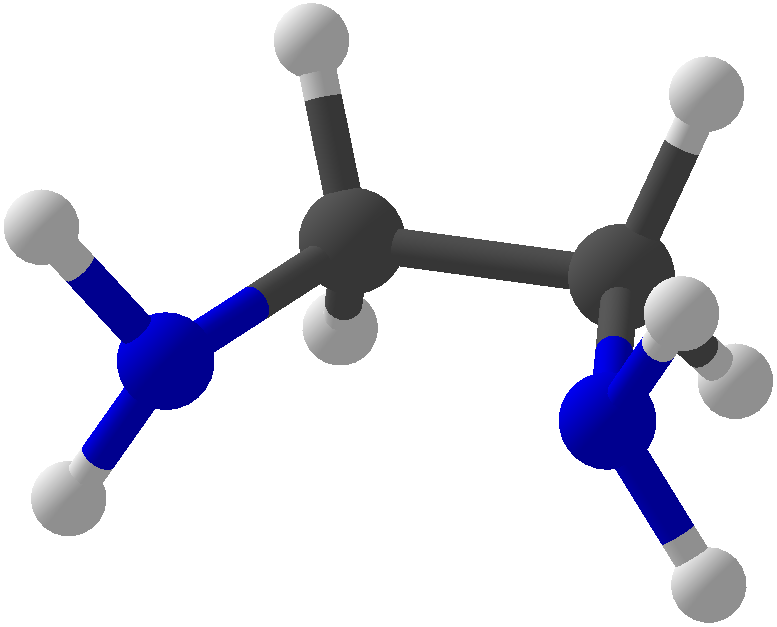
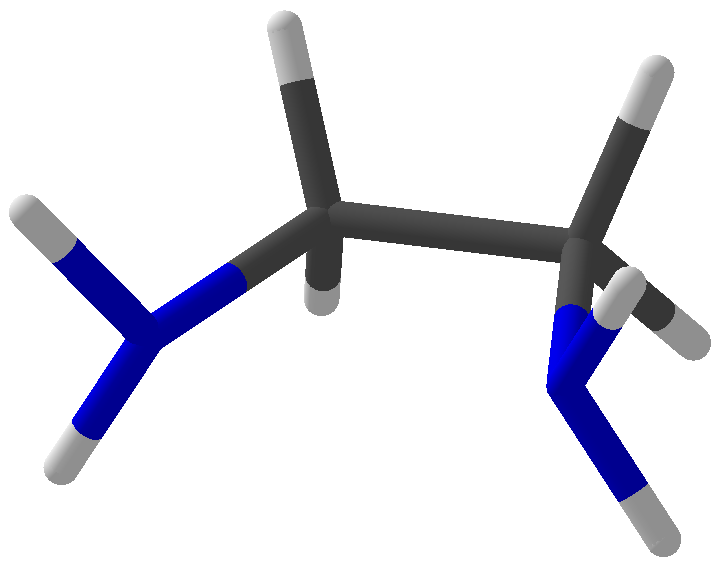
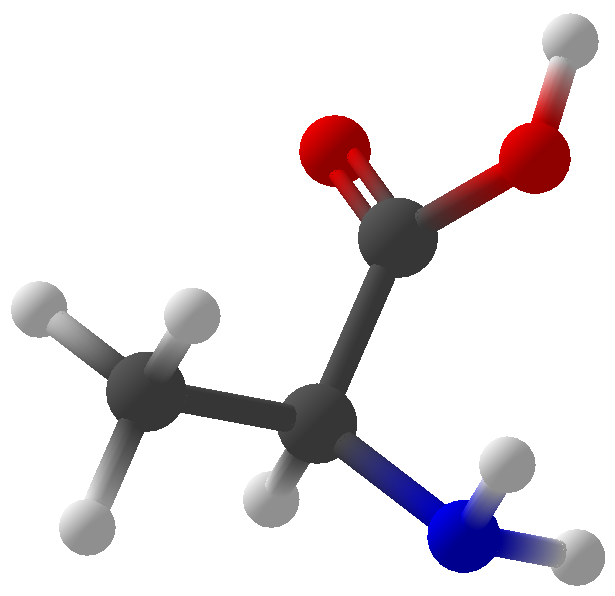
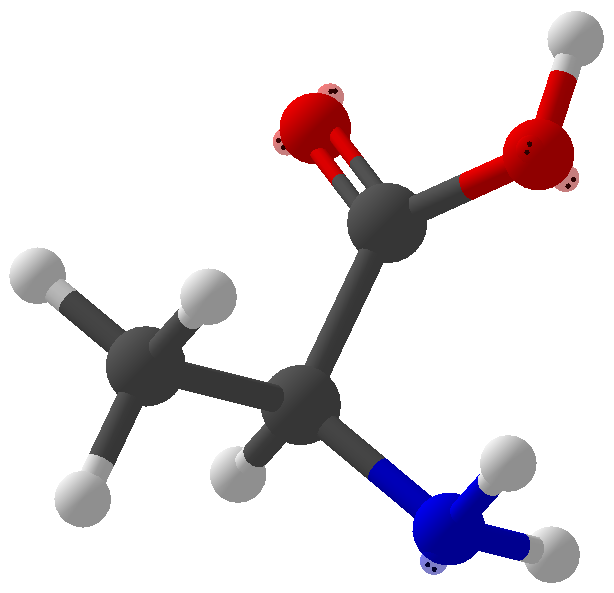
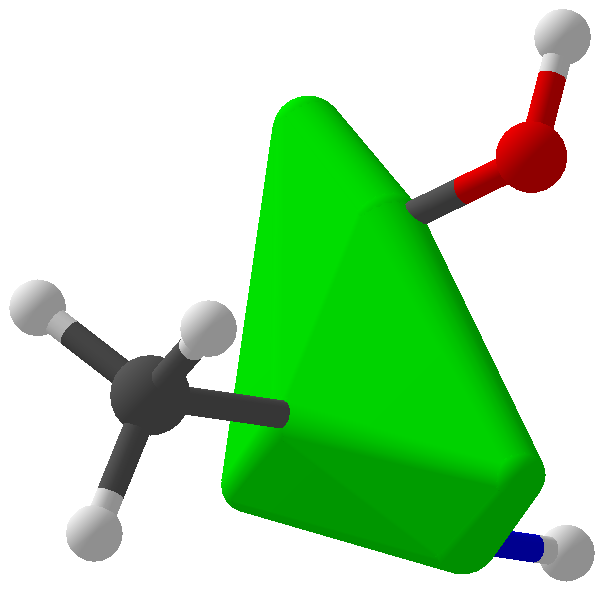
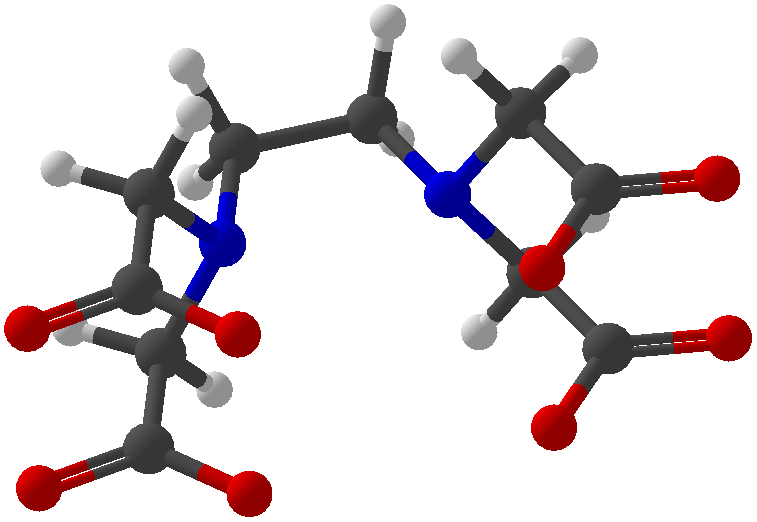
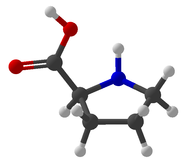
Viewing OptionsAtoms and Bonds Ball-&-stick, space-filling and skeletal models for atoms and bonds between them are common representations of molecules for display. The skeletal model is convenient for complicated molecules with many atoms. Ethyldiamine C2H4(NH2)2 Visibility The bonds are usually divided into two colors in the ratio of the respective atomic radii. If the continuous bond coloring option is chosen, the bond is colored in a smooth gradient. The "lone pairs" and "empty orbitals" can be either shown or hidden. Some molecules have precomputed convex hulls which can be displayed, lumping some atoms into a conceptual unit inside a "tight package". Alanine, one of the 20 amino acids that combine into proteins in our body Data in Color The distribution of mass, charge and electronegativity can be visualized to help understand the composition of the molecule. Each type of data is displayed as a color filter: for instance, activating the mass density filter colors each atom blue, with denser atoms in brighter blue. The other two filters operate similarly in green and red respectively, so one can discern different types of data even with overlapping colors. For example, with all three filters, a white atom has very high mass density, charge density and electronegativity.
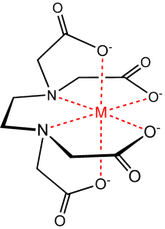
Behind the Code Hare are some of the theory and algorithms relating to chemistry and mathematics that were necessary to build this program. Refer to the source code from Github. I'll write at length in another post about the programming techniques used (mainly type classes and monads). 1 Chemistry The file Chemistry.hs contains the chemical data such as the elements up to period 4, atomic radii (Van Der Waals radii), molar masses, color designations, orbital types and geometries (based on VSEPR theory), hybridization and electronegativities. hmol can handle single, double and triple bonds between atoms, as well as "intermolecular forces" (represented as "dotted bonds" in the snapshots of DNA). Convenient subroutines build macromolecules out of atoms and smaller molecules:
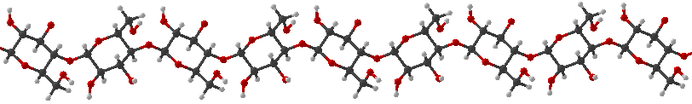
A strand of cellulose: a polymer from glucose units, it provides structural strength in plant tissue. Several strands link side-by-side to confer additional strength. I cannot vouch for the scientific accuracy of Hmol, because it was made with my high school knowledge of chemistry! The program gives a a very simplistic representation of molecules. It doesn't draw orbitals with the exotic shapes as predicted by MO theory, nor does it portray aromaticity at all. 2 Special Algorithms Algorithms of a more mathematical flavor were sometimes necessary.
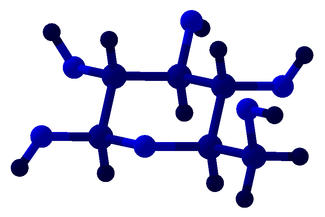
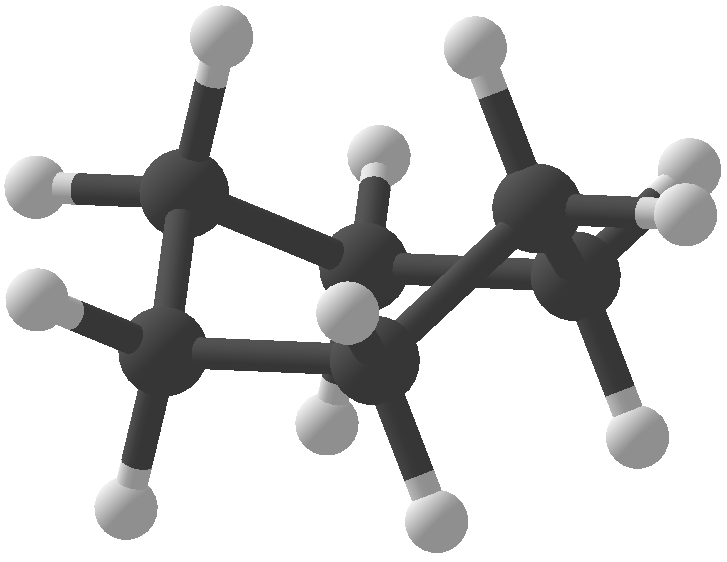
The warpAtom subroutine from Algorithms.hs fixes some orbitals of an atom in a specified configuration, then allows the remaining orbitals to roam around, "pushed" by electromagnetic repulsion from the fixed configurations. Mathematically, the repulsion between two orbitals is taken to be inverse proportional to the square of the bond angle between them. After 1000 rounds of electromagnetic jostling, the orbitals are assumed to have settled in their equilibrium positions. Is this a form of gradient descent? At any rate, the results look okay... Convex hull of spheres To compute the convex hulls of a set of atoms, normal convex hull algorithms couldn't be used because I needed the hull of spheres that could have different radii. I basically adapted the gift wrapping algorithm by considering planes that are externally tangent to three spheres at a time. The set of triangular faces is then augmented with the spheres themselves and conical frusta between adjacent spheres to render a package with a smooth surface. Cyclohexane, "boat" configuration 3 Programming The user interface was conveniently built using Glade, interfaced with Haskell via Gtk2Hs, and the 3D rendering was done using HOpenGL. Blood and sweat were shed to get HOpenGL to display what even you see in these pictures, and I'm sure that is far from the full potential of HOpenGL. I used some Haskell tricks to make programming easier, but they take long to explain so I'll save them for another post. Licensing
Copyright © 2010 Cheng Herng Yi  Graphics from Hmol in this article by Cheng Herng Yi is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. Attribution must include the Hmol program and a link to its Github repository, or if impossible, a link to this article.
1 Comment
|
Archives
December 2020
Categories
All
|


 RSS Feed
RSS Feed